Overview
The goal of this use case is to illustrate the benefit of using AgroPortal ontologies to describe, annotate and structure phenomics data within the PHIS platform, an open source information system for Plant Phenomics. We like: (i) to ease the reuse of semantic artefact objects (classes, concepts, properties, etc.) within PHIS to describe data and (ii) enable the push back of knowledge objects created by domain scientists within PHIS to application or domain ontologies hosted in AgroPortal.
Context and materials
In recent years, plant phenomics –i.e., the discipline of biology related to the measurement and analysis of the observable physical and biochemical characteristics of plants as they interact with their environment– has generated vast amounts of datasets from experiments conducted in both field and controlled conditions, encompassing hundreds of genotypes across various scales of organization. These datasets represent unprecedented resources for identifying and testing novel mechanisms and models [https://doi.org/10.1016/j.cub.2017.05.055].
However, assembling and organizing such datasets is challenging due to the heterogeneous nature of the data (e.g., environmental data, phenotypic variables, images, and metadata) and the difficulty in accessing information distributed across multiple sources.
To address these challenges, the phenomics community has proposed an ontology-driven information system, called PHIS (Phenotyping Hybrid Information System), inspired from the FAIR principles. PHIS serves as a solution for integrating, organizing, and managing multi-source and multi-scale phenomics data obtained from field and greenhouse conditions [https://doi.org/10.1111/nph.15385].
PHIS is based on the generic OpenSILEX technology, developed and maintained by INRAE-MISTEA. It is an ontology-driven open source information system tool designed for life science data. The software suite implements original management methods for the exploitation of semantics, production of FAIR data and adopts an architecture adapted to heterogeneity and increasing volumes of data. PHIS is a specific instance of the OpenSILEX technology for plant phenotyping deployed in various categories of installations (field, glasshouse) developed in part within the H2020 EPPN2020, EMPHASIS ESFRI and national PHENOME infrastructure projects.
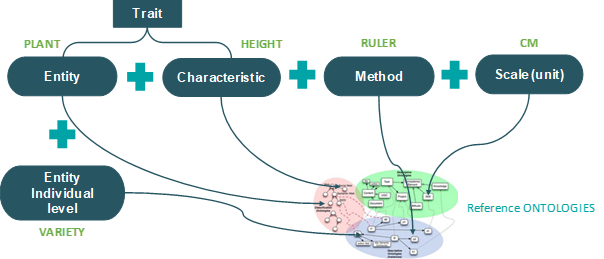
One of the major obstacles for data interoperability and reusability is the accurate identification and definition of measured variables (see the work of the I-ADOPT RDA working group). For instance, the commonly measured variable "plant height" can have various definitions depending on the crop, can be measured using different methods (e.g., image analysis or a ruler), and can be expressed in different units (e.g., cm or mm). To address this challenge, the Entity-Characteristic-Method-Unit model (https://cropontology.org/) has been adopted to facilitate the standardization of measured variables (as illustrated in Fig. 1):
- Entity: Refers to the object being targeted (e.g., plant, canopy, air, leaf).
- Characteristic: Denotes the type of measurement, encompassing physical quantities as well as observed qualities like irradiance, temperature, area, height, etc.
- Method: Describes the approach used for estimating the variable (e.g. manual measurement, image analysis, visual score).
- Unit: Describes the units used to quantify the variable (e.g., g, kg, W/m², unitless).
Within PHIS, each of these building blocks of a measured variable are mapped as much as possible to reference ontologies such as the Plant Ontology and the Crop Ontology or the SOSA ontology for sensors.
As an example, the air temperature is modeled according to this scheme as:
Air_Temperature_ShelterInstantMeasurement_DegreeCelsius
Where, the Entity is the Air, the Characteristic is the Temperature, the Method used is an instantaneous measurement using a shelter and the units are in °C.
For this variable in PHIS, the different components are mapped (when possible) to existing reference ontologies (here fetched from AgroPortal) and unique internal URIs generated by PHIS are also associated with the variable and to each of these components. In this given case, no reference term was found for the method 'ShelterInstantMeasurement,' thus the system generates a new term and associates it with an internal URI. Eventually this term will be a candidate for extending an ontology. While the former example demonstrates the flexibility and freedom of PHIS to create new terms when users are unable to find them for any reason (such as a lack of computer science skills, time constraints, or familiarity with existing repositories and resources), this approach does not promote the reuse of existing terms and limits interoperability with other resources.
- Variable URI => phis:id/variable/ev000001
- Entity (Air) => http://aims.fao.org/aos/agrovoc/c_224 (see in AgroPortal)
- Characteristic (temperature) => http://purl.obolibrary.org/obo/PATO_0000146 (see in AgroPortal)
- Method (ShelterInstantMeasurement) => phis:id/variable/method.shelter_instant
- Unit (degree Celsius) => http://purl.obolibrary.org/obo/UO_0000027 (see in AgroPortal)
This structured approach allows for the creation of new variables by combining these building blocks, for instance, by changing the method or the unit.
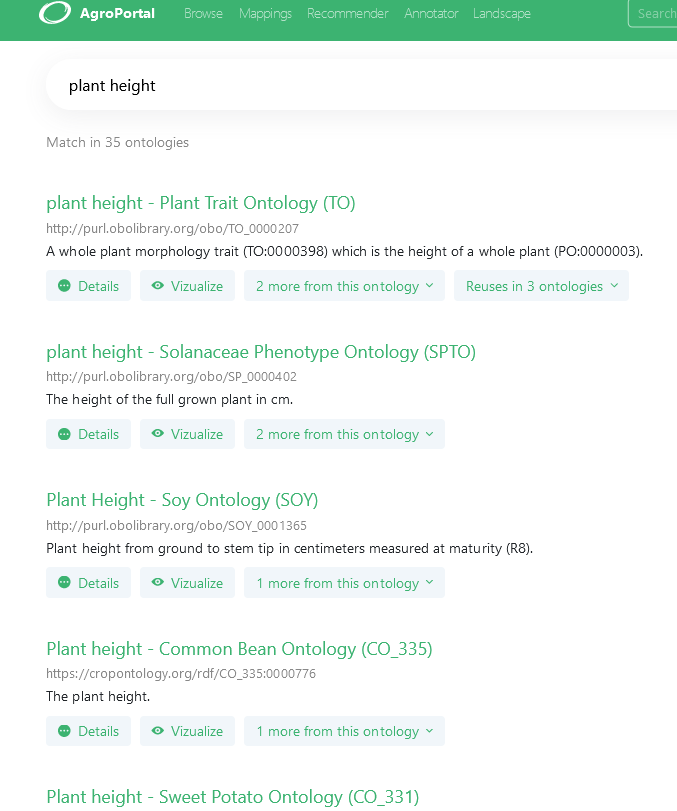
To get the relevant ontology terms for the variable, PHIS users are encouraged to use AgroPortal. It is a vocabulary and ontology repository built as a reference catalogue for hosting, sharing and serving semantic artefacts for agri-food communities, developed and maintained by INRAE-MISTEA and University of Montpellier [https://doi.org/10.1016/j.compag.2017.10.012]. AgroPortal is based on the generic technology OntoPortal developed jointly by the OntoPortal Alliance. AgroPortal allows users to search and browse for terms in a user-friendly interface (see Fig. 2). The semantic artefact catalogue can be called automatically by tools thru its API.
Challenges and objective
While the plant phenomics community has embraced ontologies to standardize the description of experimental variables, many users with a background in biology lack computer science skills and are unfamiliar with the use of ontologies or semantic artefacts, leading to difficulties in retrieving information (as illustrated in the example above). Additionally, most available resources are not centralized, which further complicates the process of gathering information from multiple sources and mapping concepts.
Currently, for PHIS users, fetching ontology terms from AgroPortal or directly from multiple sources and ad-hoc vocabulary systems is a manual process. This process requires going to another web application or tool, performing a search and manually copy/pasting the information found (if any) related to the selected ontology term. This information is then used to fill in the necessary field for the mappings by specifying the mapping (more specific, more general). This manual process considerably prevents and slows down the reuse of standard ontology terms when describing objects within PHIS.
The goal of our use case in FAIR-IMPACT T4.5 is to build a connector between PHIS and AgroPortal to ease the re-use of ontology terms when building variables and other scientific objects within PHIS.
Prototype connector between PHIS and AgroPortal
We (INRAE-MISTEA and INRAE-LEPSE) are working on a prototype (currently developed within the generic OpenSILEX technology and later moved to the PHIS instance) so PHIS users can easily describe, through Web interfaces, their measures, observations as scientific variables –using the model presented above– as well as their experimental vocabulary. The connector allows users to search and grab from AgroPortal either a term URI and its related information (name, synonyms, definition) or create a new term within PHISand describe it with information and mappings coming from AgroPortal. All the descriptions are stored in PHIS in RDF, the pivotal language of semantic knowledge graphs. Mapping between variable description and other domains ontologies are made by users with SKOS mapping relations (e.g., skos:exactMatch, skos:broadMatch).
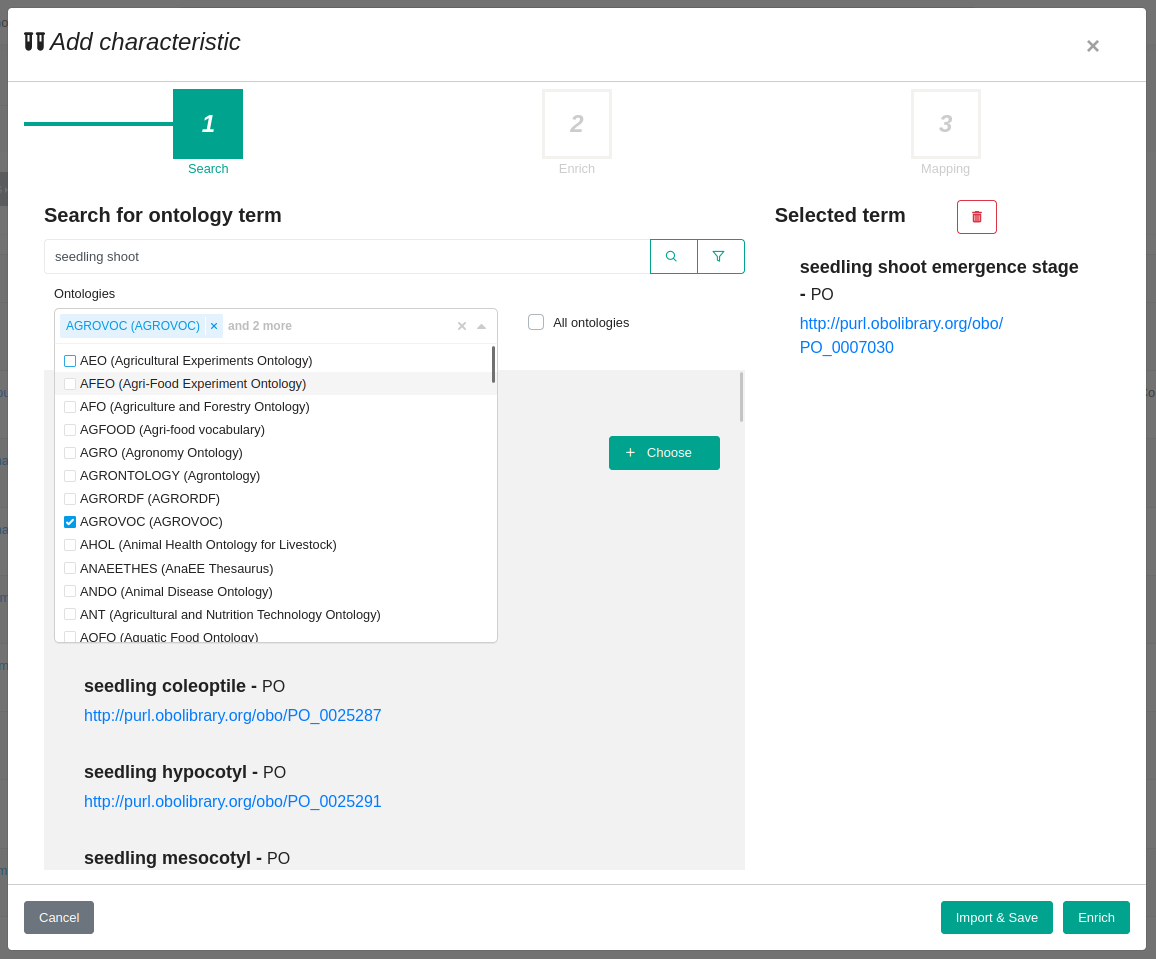
The prototype connector under development (Fig. 3) aims to address this challenge by providing a semi-automatic ontology term fetching tool embedded within PHIS. The connector is executed within an OpenSILEX instance (here PHIS) and relies on AgroPortal API to get the information. The connector will offer a number of features to make the fetching process easier and more efficient, including:
- An ergonomic search interface that allows users to easily find the terms they are looking for.
- Integration with pre-selected semantic artefacts made available by AgroPortal, such as the AGROVOC thesaurus , the Crop Ontology or multiple references ontologies for plant sciences.
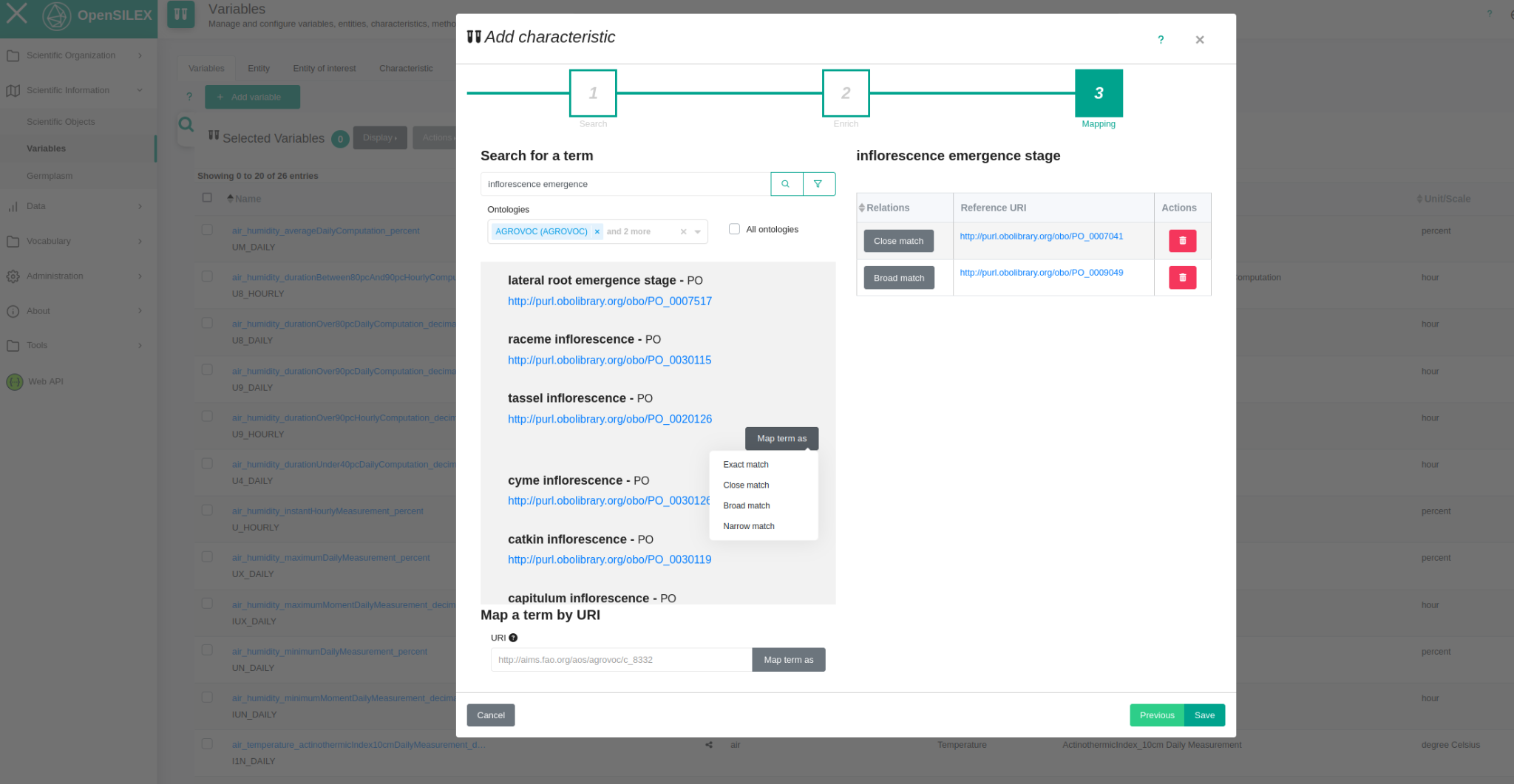
- A mapping functionality that allows users to specify the links between terms from different ontologies especially in the case a new term is created in PHIS and AgroPortal is only used to grab information and mappings to other close terms (Fig. 4).
This connector will facilitate the search for new terms to PHIS users.
In the future, following the same philosophy and technical behavior (API calls) in addition to consuming the content from AgroPortal, the connector will open up the possibility of proposing content (e.g., terms and mappings) to AgroPortal ontologies and semantic artefacts. This will valorize the new intrinsic contributions made in creating scientific objects (variables, terms, properties, etc.) by PHIS data experts which would engage with external ontology experts and users who validate these terms.
The connector will provide a number of benefits to users of PHIS and will ultimately contribute the AgroPortal’s content, including:
- Reduced time and effort required for reusing or mapping the terms used in PHIS.
- Improved interoperability between different dataset and ontologies.
The connector is currently under development as a demonstrator within FAIR-IMPACT T4.5.
Eventually, the connector would be made completely generic to work with any instance of OntoPortal (on the semantic artefact catalogue side) and on any instance of PHIS (on the data repository side).